Researchers at the Weizmann Institute of Science have developed a groundbreaking method that provides unprecedented insights into the complex relationship between bacteria and their host cells. This innovative approach is poised to revolutionize our understanding of bacterial virulence and pave the way for new strategies to tackle the growing threat of antibiotic resistance.
Dissecting the Bacterial Social Network
As it turns out, bacteria are a bit more like humans than we realize in that they do indeed have micro-social preferences and can adapt to our behaviors. There are those bacteria that prefer to navigate solitary waters, and yet there exist several others that fare well in concert with host cells. Another deadly type of bacteria, such as salmonella, are made up of a lot more cells are larger than the pneumonia bacteria and try to invade and takeover their host cells.
Earlier studies have shown the protein toxins that bacteria inject into host cells are essential. Loci for these proteins is one way that specific strains of bacteria may prove to be more infectious and disease-causing than others. These include the fact that there are over 2,500 subspecies of Salmonella, according to the researchers, but only a small number are related to life-threatening diseases. Large variation in immune responses is observed, even among individuals who carry an identical pathogen burden and this variation can mediate whether a given individual succumbs to infection or clears the bacteria.
In Paired-End Single-Cell Sequencing, A Breakthrough
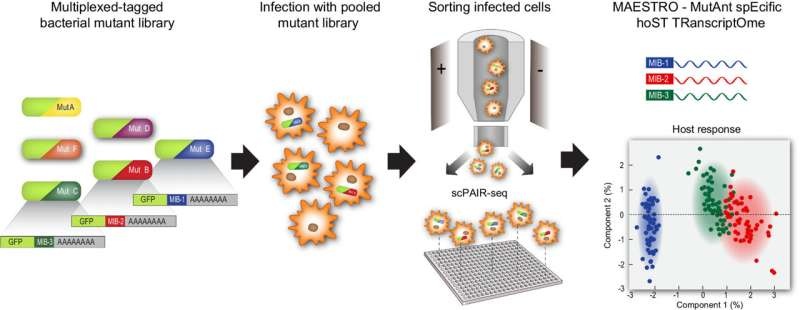
The method, developed by the Weizmann Institute crew, provides a solution to a problem that has long been plaguing microbiology. All these tools found in the public domain thus far have struggled to systematically analyze formally complete lists of host cell-bacterial subspecies interactors (due to very exciting yet difficult complex interactions with thousands and thousands of bacterial subspecies as those of your host cells). The researchers overcame this constraint with a two-step solution.
The first step was to establish a method of creating genetically engineered pools of bacterial species, each with its barcode. The researchers use the barcode DNA sequences—to identify which bacterial species is present and where it is, even inside the host cell. In the second, they built a computer model called MAESTRO that links the sequencing results of each bacterial barcode to the results of sequencing the host cell in which those bacteria have taken up residence. This “paired sequencing” method allows researchers to determine the exact influence of each bacterium on behavior and gene expression in its host cell.
Conclusion
The new system that the Weizmann Institute group has developed is considered to be ‘a game changer’ in deciphering the complex interplay between bacteria and their human host cells. This discovery should reveal novel strategies for combating the expanding threat of antibiotic resistance by increasing our understanding of how bacteria become virulent and how they interact with host immune defenses, in ways that have not been seen before. As more is learned about the functions carried out by all the proteins bacteria inject into hosts, the opportunity arises to develop new ways of treating bacterial disease, either by suppressing virulence or enhancing immune defenses in infected hosts. Such advances in paired single-cell sequencing represent a turning point in the age-old struggle against bacterial infections, bringing us one step closer to maximizing host-pathogen interactions for a future where humans and crops can be better protected from would-be infectious outsiders.
