Researchers have unraveled a key genetic mechanism behind the way pathogens infect crops, leading to new strategies for breeding resistant crop varieties against other pathogens carrying the same genetic mechanism. This discovery could pave the way for improving disease resistance in wheat, canola, and other important crops, benefiting farmers and ensuring global food security. Plant pathology and plant breeding are at the forefront of this groundbreaking research.
Unraveling the Genetic Code of Crop Diseases
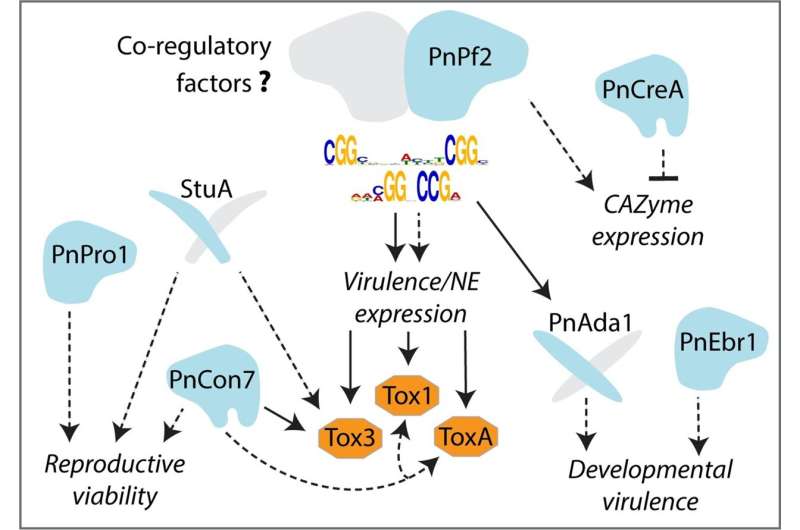
The research team, led by the Center for Crop and Disease Management (CCDM) and collaborators from Curtin University, CSIRO, the University of Nottingham, and INRAe, has made a significant breakthrough in understanding the genetic mechanism behind the way pathogens infect crops. By studying the fungus Parastagonospora nodorum, which causes septoria nodorum blotch (SNB) in wheat, the researchers were able to identify and validate the function of a specific DNA sequence linked to genes that cause damage on wheat.
The team discovered that a transcription factor called Pf2 binds to this specific DNA consensus sequence, activating adjacent genes to produce necrotrophic effectors – molecules responsible for inducing damage on wheat. This discovery is a game-changer for disease resistance breeding research, as it provides a clear understanding of how the pathogen’s effectors are activated to attack the plant.
Transferring the Genetic Insights to Other Crop Diseases
The researchers are excited about the potential applications of their findings beyond wheat. According to Associate Professor Kar-Chun Tan, the same Pf2 transcription factor is found in other fungal pathogens that cause diseases such as yellow spot of wheat, blackleg and black spot of canola. This suggests that the genetic mechanism they’ve uncovered may be a common thread in various crop diseases.
By identifying the DNA consensus sequence targeted by Pf2, the researchers can now narrow down potential effector genes and prioritize them for further investigation. This knowledge can be used as a regulatory model to accelerate the discovery of effectors in other crop pathogens, which is a crucial step towards developing more disease-resistant crop varieties.
Empowering Crop Breeders for a Sustainable Future
The findings of this research have far-reaching implications for the future of crop production and food security. CCDM Director Professor Mark Gibberd expressed his pride in the team’s perseverance and their ability to solve a complex scientific mystery that will lead to better crop varieties for growers.
With the discovery of the DNA consensus sequence, crop breeders can now use effector-assisted selection to identify and breed for crops with improved disease resistance. This knowledge can be applied not only to wheat but also to other important crops like canola, ensuring that farmers have access to more resilient and productive varieties. By unlocking the genetic secrets behind crop disease infection, this research paves the way for a more sustainable and food-secure future for agriculture.
