A groundbreaking study published in Nature Communications presents an AI-driven approach that upends traditional views on protein structure relationships. Led by a team from the University of Virginia, this research leverages deep learning and a novel conceptual model, the Urfold, to uncover previously overlooked connections across the protein universe.
Revolutionizing Protein Structure Analysis
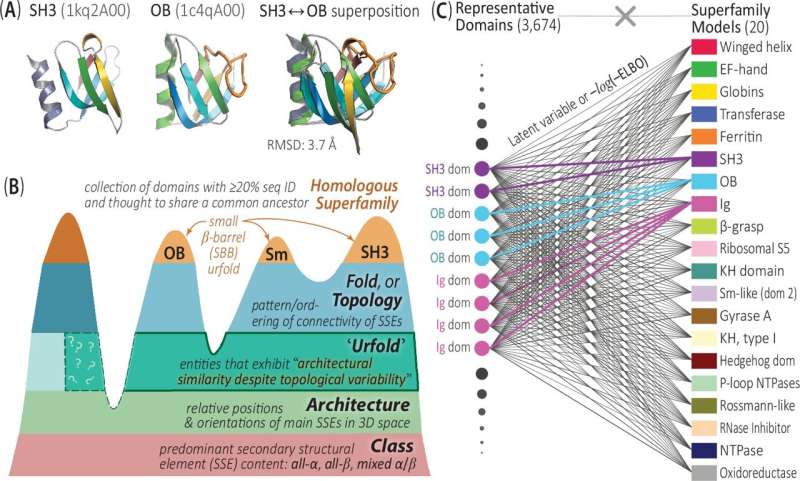
The traditional approach to classifying protein structures has often relied on rigid, non-overlapping categories. However, the research team from the University of Virginia has developed a groundbreaking AI-driven framework called DeepUrfold that challenges this conventional wisdom.
Using deep learning techniques, DeepUrfold is able to detect and quantify subtle structural relationships between proteins that would otherwise be missed by traditional methods. By considering “communities” of proteins rather than rigid bins, the researchers have uncovered a more nuanced and flexible understanding of protein structure similarities and differences.
Uncovering Distant Structural Relationships
A key aspect of the study is the introduction of the Urfold, a new conceptual model that allows for proteins to exhibit architectural similarity despite having different topologies or “folds.” This innovative approach has enabled the research team to identify numerous faint structural relationships across the protein universe that were previously undetected.
By leveraging the power of AI and the Urfold model, the researchers have been able to capture and describe these distant relationships, pushing the boundaries of our understanding of protein structure and evolution. This shift in perspective could have far-reaching implications for fields such as structural biology, computational biology, and drug discovery.
Empowering a Holistic Approach to Protein Studies
The findings of this study challenge researchers to move beyond thinking of protein similarities in purely geometric terms and to adopt a more integrated, dynamic approach. By embracing the Urfold model and the insights provided by DeepUrfold, scientists can now explore protein relationships in a more nuanced and comprehensive manner.
This shift in perspective could inspire new avenues of research and collaboration, as scientists work to unravel the complex web of protein structure and function. As the field continues to evolve, the work of the University of Virginia team stands as a testament to the transformative power of AI-driven approaches in advancing our understanding of the protein universe.
