Researchers at Waseda University, in collaboration with bitBiome, Inc., have developed a groundbreaking single-cell genome approach that is shedding new light on the human microbiome and its role in antibiotic resistance. This innovative technique has the potential to transform how we understand and combat the spread of antibiotic-resistant bacteria, with far-reaching implications for public health, environmental monitoring, and agriculture. Microbiome research has never been more critical, and this study showcases the remarkable power of single-cell genomics in unlocking the secrets of microbial diversity and interactions.
Revolutionizing Microbiome Research with Single-Cell Genomics
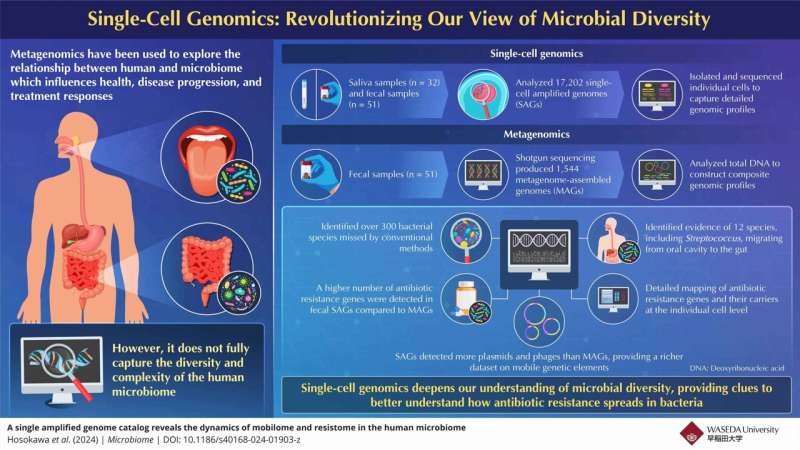
The human microbiome is a complex and dynamic ecosystem that plays a crucial role in our overall health and well-being. Traditional metagenomic approaches have provided valuable insights, but they often fall short in resolving microbial diversity at the strain level and accurately profiling genes involved in antibiotic resistance.
To address these limitations, a team of researchers led by Associate Professor Masahito Hosokawa from Waseda University, in collaboration with bitBiome, Inc., developed a groundbreaking single-cell genome approach. This innovative technique allows for the detailed analysis of individual bacterial cells, providing a more comprehensive understanding of microbial communities and their genetic features, including antibiotic resistance genes.
The researchers analyzed over 30,000 individual genomes of oral and intestinal bacteria, the largest dataset of its kind, showcasing the remarkable potential of single-cell genomics in elucidating microbial diversity and interactions.
Uncovering the Secrets of Antibiotic Resistance in the Human Microbiome
One of the key findings from the study is the in-depth profiling of antibiotic resistance genes within the human microbiome. By examining individual bacterial cells, the researchers were able to gain deeper insights into how these genes are exchanged and spread, a critical factor in the growing threat of antibiotic resistance.
The detailed mapping of antibiotic resistance genes can have far-reaching implications for public health. This information can inform the development of more targeted and effective treatment strategies, helping to prevent diseases, reduce healthcare costs, and improve overall public health outcomes.
Beyond the medical realm, the single-cell genomics approach can also aid in environmental monitoring and management. By tracking genetic shifts in microbial communities across different ecosystems, scientists can better understand and mitigate the spread of antibiotic resistance, a pressing global concern.
Advancing Agriculture and Sustainability with Single-Cell Genomics
The potential applications of this single-cell genomics technology extend beyond the medical field and into the agricultural sector as well. By understanding the dynamics of antibiotic resistance gene exchange within microbial communities, researchers can guide farming practices to minimize the spread of resistance through soil, water, and livestock.
This knowledge can lead to more sustainable and responsible agricultural practices, reducing the reliance on antibiotics and mitigating the risk of antibiotic-resistant pathogens emerging in our food supply. Furthermore, this approach can provide valuable insights into the role of the microbiome in plant and animal health, potentially leading to innovative solutions for improving agricultural productivity and sustainability.
The transformative power of single-cell genomics is undeniable, as this study demonstrates. By unlocking the secrets of the human microbiome and antibiotic resistance, researchers are paving the way for groundbreaking advancements in public health, environmental protection, and sustainable agriculture, ultimately benefiting both human and planetary well-being.
