Researchers have uncovered a fascinating world of commensal Neisseria bacteria living in the human oropharynx (throat). These microbes, while harmless to us, possess a remarkable ability to share antimicrobial resistance genes with their more notorious cousins, the pathogenic Neisseria gonorrhoeae and Neisseria meningitidis. This study provides critical insights into the prevalence, diversity, and antimicrobial resistance profiles of these commensal Neisseria species, highlighting their potential role as a reservoir for the spread of antibiotic resistance. The findings have significant implications for understanding and combating the growing threat of antimicrobial resistance in the years to come.
Commensal Neisseria: Unsung Heroes of the Oropharynx
The human oropharynx, the area at the back of the throat, is home to a diverse community of microorganisms, including a group of bacteria known as commensal Neisseria. These bacteria are considered “commensal” because they typically do not cause disease in healthy individuals. However, their role in the broader ecosystem of the human body is far from trivial.
Uncovering the Prevalence and Diversity of Commensal Neisseria
In this study, researchers from the London School of Hygiene & Tropical Medicine set out to investigate the carriage and antimicrobial susceptibility of commensal Neisseria species in the oropharynx of 50 participants. Their findings were remarkable: 86% of the participants were colonized with at least one Neisseria species, and 66% were harboring more than one isolate.
The most common species identified was Neisseria subflava, accounting for 61.4% of the isolates. Other species, such as Neisseria flavescens, Neisseria perflava, Neisseria macacae, and Neisseria mucosa, were also present in varying proportions.

Antimicrobial Resistance: A Concerning Trend
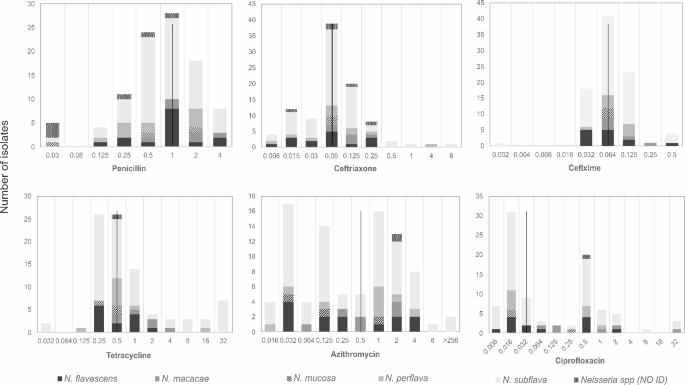
The researchers didn’t just stop at identifying the Neisseria species; they also investigated their antimicrobial susceptibility profiles. This is where the true significance of their findings comes into play.
Alarmingly, the team found that the resistance rates for certain antibiotics, such as ceftriaxone (a crucial cephalosporin antibiotic) and azithromycin (a macrolide antibiotic), were above the World Health Organization’s (WHO) threshold of 5% for changing the first-line treatment. Specifically, the resistance rates were 5% (according to CLSI guidelines) and 13% (according to EUCAST guidelines) for ceftriaxone, and 29.3% for azithromycin.
Genetic Insights: Uncovering the Resistance Arsenal
To better understand the genetic basis of this antimicrobial resistance, the researchers performed whole-genome sequencing on 30 of the Neisseria isolates. Their analysis revealed the presence of several antimicrobial resistance genes, including those conferring resistance to macrolides and tetracyclines.
Interestingly, the team also identified the presence of DNA uptake sequences (DUS) in the genomes of the commensal Neisseria species. These sequences play a crucial role in facilitating the gonorrhoeae’>Neisseria gonorrhoeae and resistance’>antimicrobial resistance.
Conclusion: Uncovering the Microbial Secrets of the Oropharynx
This study has shone a spotlight on the fascinating and complex world of commensal Neisseria bacteria in the human oropharynx. By unveiling their prevalence, diversity, and antimicrobial resistance profiles, the researchers have opened up new avenues for understanding and addressing the global challenge of antimicrobial resistance. As we continue to unravel the secrets of the human microbiome, studies like this one will undoubtedly play a crucial role in our efforts to safeguard public health and ensure the continued effectiveness of our antimicrobial arsenal.
Author credit: This article is based on research by Victoria F. Miari, Wesley Bonnin, Imogen K. G. Smith, Megan F. Horney, Samer J. Saint-Geris, Richard A. Stabler.
For More Related Articles Click Here
