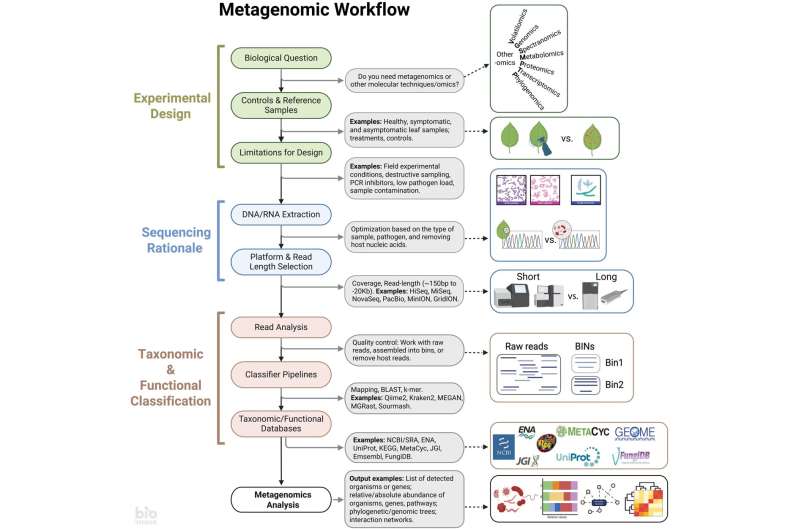
Despite the vast number of microbes on Earth, only a tiny fraction have been discovered. A new approach called metagenomics may hold the key to unveiling these elusive organisms and their role in plant health. This innovative DNA sequencing technique allows researchers to analyze entire microbial communities, offering unprecedented insights into the complex interactions between plants, pathogens, and the broader microbial ecosystem.
Making Visible the Invisible Accomplices
Because plants are so full of life — there can be as many as 100 million bacterial cells in a gram of leaf matter and 1 trillion bacteria in the soil around the roots. Microbial populations can have drastic effects on plant health, either by supporting or inhibiting a specific plant’s capacity to thrive. Nevertheless, the overwhelming majority of these microbes have been virtually invisible and unknown before this discovery because they have defied cultivation in the laboratory.
Now, venturing into the metagenomic territory, Penn State researchers have devised a novel strategy to unmask the secrets of these missing microbes. They analyze genetic material from environmental samples — say, soil or leaves or even air itself — to see what microbes are present and how they differ in kind and function. Such an approach has been extremely useful for biologists, who typically study all the species of microorganisms present (as opposed to just handfuls at a time) to get a broader sense of what goes on with these microscopic communities without engaging in traditional isolation and culturing methods which are often not comprehensive.
Microbial Coexistence and Pathogen Control
A greater insight into the interactions between pathogens and the rest of the plant-associated microbial community is critical in plant pathology. Plant pathogens like fungi can result in crop losses of up to 20%, making them the single biggest threat to global food security. Yet it is not a one-way interaction, because the relationship between pathogens and the microbiome of the plant can make disease worse or milder.
Because metagenomics enables for the first time a detailed view of the complete range of pathogens, researchers can study pathogen diversity related to population movement and compare bacterial populations in different areas with similar disease outbreaks. This action additionally permits quite brisk acknowledgment of a pathogen that is very hard to develop, offering an unbiased technique for distinguishing all likely ailment-creating specialists. Insights into these webs of interactions help researchers uncover roots (pun intended) of plant disease, and better inform strategies to help farmers keep crops healthy.
Unraveling mysteries of plant-microbe symbiosis
In addition to disease diagnostics, metagenomics provides information on the overall microbial ecology in a plant system. Researchers can discover how these organisms interact with one another, share genes, and respond to changes in their surroundings by sequencing the genetic material of the entire microbial community. This involves studying antibiotic resistance, the evolution of new pathogenic species, and the traits behind how well-known microbes have adapted to their plant hosts.
These findings can help guide attempts to cultivate a plant microbiome in balance with its host and ideally be able to trigger an improved immune response in the plant’s defense against pathogens. Through the use of metagenomics, researchers can obtain a more detailed view of the complex network of processes that maintain the plant’s microbiome which could open new routes in breeding microbes for agriculture and food security.
