Researchers from the NUS Institute for Health Innovation & Technology have developed a groundbreaking technology called TETRIS that uses DNA barcodes to map complex protein interactions within tumor cells. This innovation promises to revolutionize cancer diagnostics by enabling more precise subtyping and identification of aggressive forms of the disease in just a few hours. Unlike current methods, TETRIS can capture the comprehensive hierarchy of protein interactions, providing vital insights for tailoring personalized treatment strategies.
Unraveling the Protein Puzzle
Proteins, on the other hand, are located at almost the base level of any process. These proteins interact with each other, a phenomenon that is essential for biology and medicine.
Yeast-two hybrid assays and mass spectrometry-based proteomics are just two of the current methods available, but they too have limitations in identifying protein interactions because they may not be capturing all possible types of interactions (particularly higher-order ones) that often contribute to the most aggressive clinically relevant cancer phenotypes.
So the NUS researchers said current methods lack a complete nature and potentially correct approach for studying protein interactions. To overcome these limitations, the researchers turned to DNA nanotechnology–a field that allows precise programmability and predictable interactions of molecules.
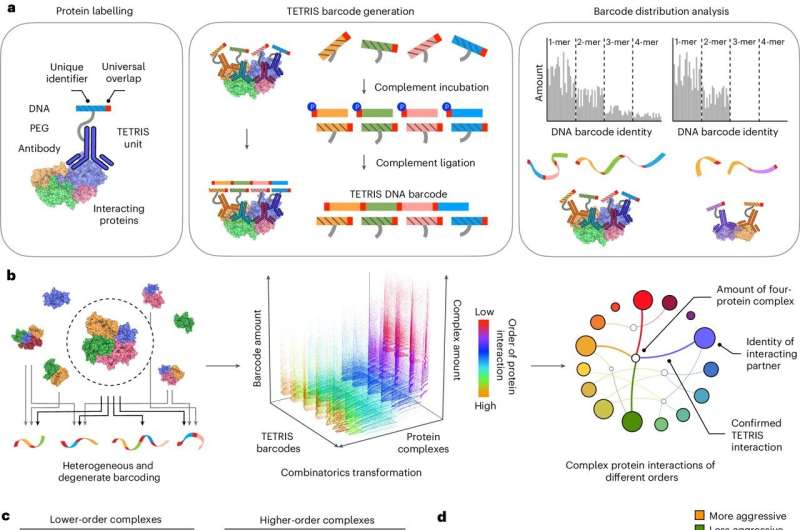
TETRIS: The Social Networking of Proteins
With its unique ability to produce hybrid molecular structures acting as smart encoders, the TETRIS technology can be used for identification of protein interfaces directly within patient samples. Each rapporter encoder introduces a target-recognizing antibody and a template DNA barcode. The encoders are fused bilaterally when they bind to interacting proteins and the barcode captures both the molecular identity and the spatial relationship of these proteins.
TETRIS can assess not only pairwise but also higher-order protein interactions and therefore add an important layer of complexity to the intricate protein interactome, which has been missing from available methodologies to date. This provides a more comprehensive insight into the underlying molecular mechanisms aiding disease progression that can help in better categorizing cancer and develop personalized treatment protocol.
Proteins are like the delegates at a scientific conference. Delegates each have a barcode on their name tag. Professor Brian Lim, who developed the algorithms used to process TETRIS’s data said: “When they interact or ‘shake hands’ with one another, TETRIS is able to record that interaction by chaining their barcodes together.
Revolutionizing Cancer Diagnostics and Therapeutics
In biopsies of human breast cancer tissues, TETRIS shows the ability to diagnosing cancer subtypes accurately and estimating higher-order protein interactions that are associated with aggressiveness in cancers. Since that time, this technology has transformed our understanding of diseases at the molecular level–opening a new field (transcriptomics) and revealing countless mysteries about all kinds of diseases including cancer.
Families that detect changes in higher-level protein interactions–a hallmark of the aggressive cancer disease state–could help drive more informed and personalized clinical decisions. Moreover, the technology is meant to be scalable and flexible for easy integration with regular clinical workflows.
By using TETRIS for different types of cancers and neurological diseases, the team of researchers at NUS hope to leverage it as a powered-up diagnostic tools and therapeutic interventions development platform that could work on various illnesses. In the near future, given these two filed patents and a team with plans to bring the innovation to market, there may be TETRIS within sight in how we combat disease.
