Researchers have developed a novel sequencing method that provides an unprecedented glimpse into the complex and intricate relationship between bacteria and their host cells. This revolutionary approach sheds light on the virulence factors that make some bacterial subspecies more deadly than others, paving the way for new strategies to combat antibiotic resistance.
Unraveling the Bacterial Social Network
The authors note that bacteria, like humans, have their social preferences. Some are fastidiously free form, while others revel in a host cell. Salmonella — and many of its bacterial siblings — fall into the latter, where a protein is literally injected, hijacking the host cell’s systems.
In the last few years, scientists have described how different bacterial subspecies deliver assorted syringe proteins to target cells; those differences might help explain why some are more virulent than others. For instance, there are more than 2,500 varieties of Salmonella, yet just some people cause serious infections. A new research method developed by Prof. Roi Avraham and his team at the Weizmann Institute of Science will attempt to close these gaps with some much-needed information.
Revolutionizing Host-Pathogen Interaction Research
Recent advances in molecular biology, most notably single cell sequencing techniques, have completely changed how researchers can study the complex bacterial/host interactions. However, the abilities of current tools are limited in guise of these interactions to be realized at their full scale — especially when finding with thousands of bacterial subspecies and an extensive diversity in host cells.
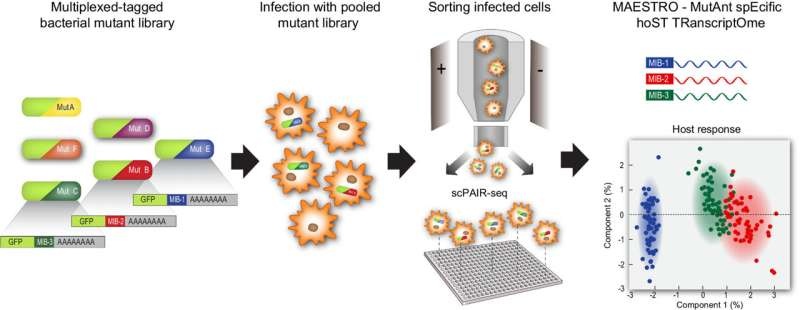
This is exactly the issue Avraham’s team has found a new way to address in two critical ways. The approach takes advantage of a way, developed by Dr. Ori Heyman, for bar coding different bacteria, which makes it possible to follow each single bacterium even as it resides within the host cell. The sequencing results of each bacterial barcode are matched by the computer model, MAESTRO, developed by Dr. Noa Bossel Ben-Moshe, to those of the specific host cell it has infected.
Detailed analysis of how the behavior of each mutant bacterial species influences that of the host and which proteins are uniquely expressed in infected host cells requires ‘paired sequencing’ at the single-cell level between bacteria and host. This level of granularity has never before been achieved, which enables a new understanding of the host-pathogen interplay.
Conclusion
The method of sequencing developed by the research team at the Weizmann Institute of Scienceborders on revolutionary and can open up a whole new way that we see the relationship that exists between bacteria and their hosts. This could provide new strategies to tackle one of the most important global challenges in health today: antibiotic resistance by revealing what key virulence factors make some bacterial subspecies more lethal than others. Given the complexity of these host-pathogen interactions and research into them, the opportunities for both basic science and applications to advance are really pretty thrilling.
