Discover how a new AI tool, PertKGE, is revolutionizing drug discovery by efficiently deconvoluting the complex interactions between compounds and proteins, paving the way for accelerated drug development.
Breaking HighThroughput Barriers in Drug Discovery
The basic components of drug discovery compounds and proteins are simple, but the intricate interplay between the two is not easy to capture. Over recent years all compound-protein interactions (CPIs) cannot be predicted and explained accurately by traditional computational methods.
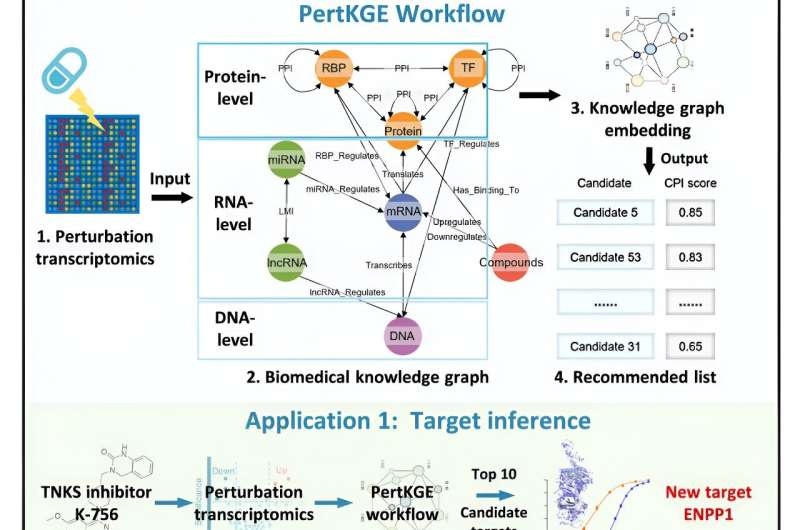
Life Within a Novel AI Advice Tool for Drug DiscoveryNeutral Editing —> A recent game-changer in drug discovery by means of synthetic intelligence (AI) is an AI device referred to as PertKGE. PertKGE was developed by a research team of the Shanghai Institute of Materia Medica (SIMM)of Chinese Academy of Sciences to deconvolute CPIs and integrate perturbation transcriptomics with knowledge graph embedding.
Here we present a novel transcriptomics concept in drug discovery — Perturbation Transcriptomics that places relevant entities (compounds) next to their potential effects at gene expression level. This approach directly reports on the molecular basis of how chemical leads perturb cellular systems, both individual cells and cell lines (or even patients), which in turn provides a window into underlying CPI mechanisms.
Unique way of PertKGE
Unlike the majority of classical methods, PertKGE builds on top of the widely-used prior biomedical knowledge graphs but it does so in an entirely new way. The main innovation is building a biologically relevant knowledge graph exploring the nitty-gritty of gene regulation.
PertKGE will thus take genes all the way down to the nitty-gritty details that ultimately cause a gene’s presence or absence on disease-gene prediction lists, including DNA, mRNA, lncRNA, miRNA, TFs and RBPs. This holistic method empowers PertKGE to cover multiple granular interactions among these regulators events, which is critical in imitating the intricate post-transcriptional and post-translational incidents that occur inside biological organisms.
With the DFS setting and DistMult embeddings, PertKGE can map all of these modules into a deeply informative hidden space to subsequent decintegrate yse CPI from perturbation transcriptomics data.
Conclusion
Well, the emerge of PertKGE is a great advancement in drug discovery. This cutting-edge AI tool has the promise to speed up the discovery of novel drug targets and improve therapy development by integrating perturbation transcriptomics with knowledge graph embedding. With the refined as well as the consolidated regulatory information in PertKGE, we presume the predictions of both performance and application breadth will be enhanced further along with facilitating an increasing influence on drug discovery.
