Researchers at the Institute for Research in Biomedicine (IRB) Barcelona have developed a revolutionary computational technique that can accurately predict gene architecture by analyzing nucleosome positions. This innovative approach, which combines experimental methods and machine learning, offers new insights into the complex interplay between DNA packaging and gene expression. DNA and nucleosomes play a crucial role in the efficient functioning of living organisms, and this study paves the way for a deeper understanding of these fundamental biological processes.
Decoding the Cryptic Art of DNA Packaging
DNA, a molecule embedded in the 46 chromosomes of human cells, is stored as very structured sculptural folds so that it can be read and processed correctly. This intricate process is mediated by nucleosomes which are key players involved in DNA compaction and gene expression regulation.
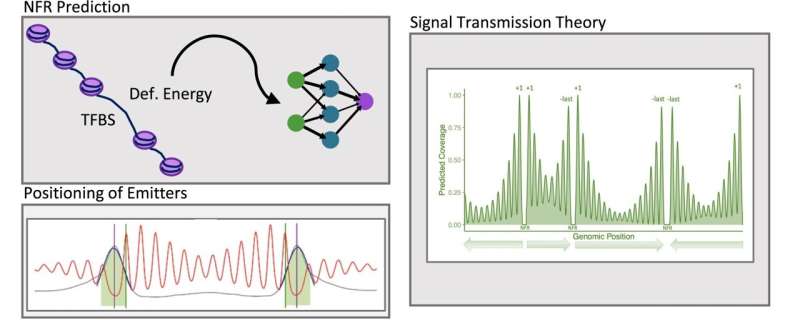
Now researchers under Dr. Modesto Orozco’s leadership––at IRB Barcelona has discovered a ground-breaking technique, Bioinformatics Prediction of Nucleosome Positioning and Occupancy (BiPNP), to predict the exact sites in which nucleosomes are responsible for DNA folding efforts. With the mixture of experimental techniques such as MNase-seq, machine learning and signal transduction theory, our strategy enables to not only replicate existing experimental data but also predict nucleosome positions faster and more accurately than ever.
Mapping DNA Sequence and Physical Signals
The IRB Barcelona team was able to show that there are two forces, the DNA sequence and physical signals released by gene ends, that shape the architecture of nucleosomes. They define the positions of the first and last nucleosomes (+1, -last) and also affect nucleosome positioning across genes.
Nucleosome structure “may affect gene expression in ways we thought were more simple,” says Ph.Alba Sala ‘researcher’, who led the study. D. candidate at IRB Barcelona and first author of the study Besides, this discovery is revolutionary and could change our knowledge about how chromatin structure can drive disease pathogenesis.
Creating New Roads for Better Therapeutic Opportunities
An improved understanding of DNA and nucleosome organization is likely essential to discover new therapeutic targets and develop more effective treatments. As a tool, this computational method represents an important advance in the broader field of epigenetics that investigates how alterations to chromatin structure globally can impact gene expression and lead to a variety of health problems.
According to Dr. Orozco, head of the Molecular Modelling and Bioinformatics lab at IRB Barcelona and Full Professor at the University of Barcelona, “Our model is as accurate as back-of-the-envelope calculations go for experimental systems. This work represents one of the most exciting milestones in our previous understanding of gene expression and will undoubtedly serve as a fundamental whetstone to allow for future investigations into how DNA packaging influences the causes of diseases. Now that you have this new toy, you can really start digging into the tiny details of how life works.
