Cutting-edge research has unveiled a groundbreaking single-cell genomics approach that is transforming our understanding of the human microbiome and its role in antibiotic resistance. This study, led by Associate Professor Masahito Hosokawa from Waseda University, showcases the power of this innovative technique in unraveling the intricate dynamics of microbial communities and their genetic features. By delving into the world’s largest genome dataset, the researchers have paved the way for advancements in public health, environmental monitoring, and the agricultural sector. Human Microbiome Antibiotic Resistance
Revolutionizing Microbial Diversity Analysis
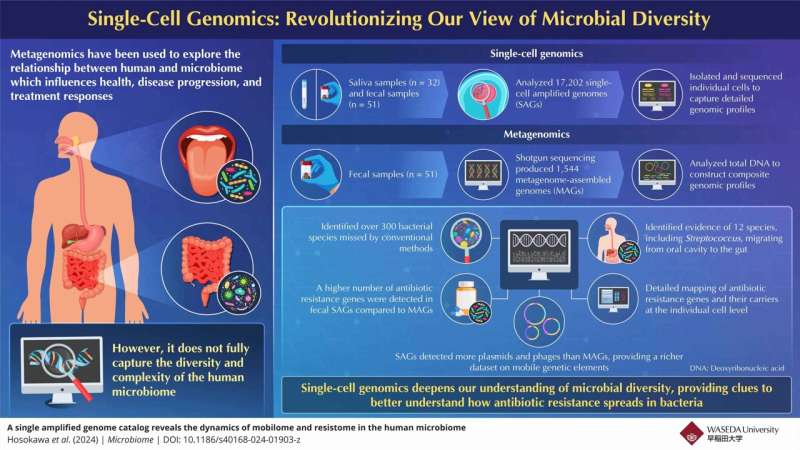
Although conventional metagenomics has enabled important insights into the human microbiome, it has limitations in the resolution of microbial diversity at the strain-level and functional gene profiling for antibiotic resistance. This shortcoming created the motivation and drive for a more sophisticated method, inspiring researchers to create an innovative single-cell genome-based strategy.
The single-cell genome approach, implemented in the commercial bit-MAP platform by bitBiome Inc., stands out as a complementary solution to conventional metagenomics. With this method, the researchers analyzed single bacterial cells and were able to recover genomes of 300 bacterial species that had been overlooked previously. It opened up a new level of resolution for probing complex networks of antibiotic resistance genes, bacterial interactions, and microbial diversity on an unprecedented scale.
Unlocking the Secrets of Antibiotic Resistance
One of the study’s tools was a single-cell genomic analysis that revealed details about changes in antibiotic resistance throughout the human microbiome. The researchers analyzed 30,000 individual genomes of oral and gut bacterial species, painting a comprehensive picture of how hundreds of the bacteria heaja’s most innocuous share genes — including antibiotic-resistance genes.
These deep profiles of antibiotic-resistance genes have broad implications for public health. These insights can be critical for designing better, more tailored treatments that prevent disease, save money, and preserve public health. Finally, by employing single-cel genomics techniques, this approach has the potential to track genetic changes in ecosystems over time which could potentially be used to monitor and minimise the spread of antibiotic resistance in the environment and agricultural settings.
Transformative Potential of Single-Cell Genomics
The research team led by Associate Professor Masahito Hosokawa and others has done the first work of this kind, a groundbreaking result that revealed the power of single-cell genomics as a tool for understanding microbiomes. With the analysis of this largest genome dataset found on Earth, they have shown that microbial communities and their interactions are not as clear-cut.
The approach offers great potential for application in subsequent medical and public health-related studies. Such studies of the complicated webs that drive drug resistance can provide important tools for other tools for promising to control this pressing worldwide health crisis. The knowledge obtained from this study can also inform environmental monitoring and agricultural practices that decrease the dissemination of antibiotic resistance and promote sustainability.
